Single-Cell CUT&Tag: Sensitive Chromatin Profiling at Single-Cell Resolution
Single-cell CUT&Tag offers targeted, sensitive chromatin profiling in individual cells with minimal input and streamlined workflows. By measuring histone modifications and transcription factor binding directly in intact cells, scCUT&Tag captures rich regulatory information across complex tissues and states including rare and transitional populations while reducing sequencing costs and experimental complexity.
What Is Single-Cell CUT&Tag?
Single-cell CUT&Tag (scCUT&Tag) is an advanced method that couples antibody-targeted in situ chromatin cleavage with droplet or nanowell barcoding to profile histone modifications and transcription factor binding in individual cells. By performing the CUT&Tag chemistry directly in intact cells or nuclei and integrating with single-cell platforms, scCUT&Tag delivers epigenetic landscapes at single-cell resolution with exceptional sensitivity and minimal background.
Core Innovation
Unlike traditional ChIP-seq approaches that require cell pooling and extensive handling, scCUT&Tag uses in situ tagmentation to tag DNA directly at antibody-bound chromatin sites within individual cells. This minimizes DNA loss, simplifies the workflow, and enables seamless integration with droplet and nanowell platforms, delivering high-quality data from tens of thousands of cells per experiment while dramatically reducing sequencing costs.
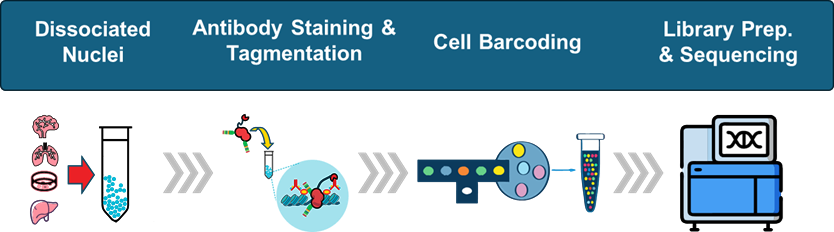

How Single-Cell CUT&Tag Works
-
Cell Permeabilization
Cells or nuclei are gently permeabilized to allow antibody access to chromatin while maintaining cellular integrity and spatial organization.
-
Antibody Binding
Antibodies specific to histone marks, transcription factors, or other chromatin-associated proteins bind to their cognate targets in chromatin.
-
In Situ Tagmentation
Protein A-Tn5 fusion directs transposase to antibody-bound sites, inserting sequencing adapters directly into DNA at chromatin target regions without requiring enzymatic cleavage.
-
Single-Cell Barcoding
Tagged nuclei are encapsulated with 10x Chromium, Illumina PIPSeq, BD Rhapsody, or other platforms. Each cell receives a unique barcode linking all its chromatin fragments.
-
Library Preparation & Sequencing
Libraries are generated using standard protocols, sequenced, and analyzed to produce cell-by-peak matrices revealing histone mark or TF landscapes in individual cells.
Platform Compatibility
Single-Cell CUT&Tag kits from Epigenome Technologies are compatible with a variety of different single-cell platforms, giving you flexibility and choice in your experimental scale and throughput.
- 10x Genomics Chromium: Direct integration with standard droplet workflows
- Illumina PIPSeq: Emulsion-based high-throughput profiling
- BD Rhapsody: Microwell-based high-throughput profiling
- Spatial implementations: Takara Trekker spatial barcoding
Need assistance? Speak with a scientist to discuss platform selection and optimization.
Strengths and Capabilities of Single-Cell CUT&Tag
1. High Sensitivity and Low Background
scCUT&Tag generates high-quality data with exceptional signal-to-noise. Targeted in situ tagmentation eliminates non-specific background inherent to immunoprecipitation-based methods, resulting in precise chromatin maps with far fewer cells and substantially lower sequencing depth than ChIP-seq.
- Precise: Direct tagmentation at antibody-bound sites minimizes off-target noise
- Efficient: Same sequencing depth requirements as scATAC-seq
- Reliable: Detection of weak or transient chromatin marks
2. Ultra-Low-Input and Streamlined Workflow
The in situ chemistry is performed directly in intact cells or nuclei without cross-linking or extensive handling steps. This minimizes DNA loss, simplifies sample preparation, and makes scCUT&Tag inherently compatible with droplet and nanowell barcoding platforms.
- Streamlined: Minimal sample manipulation and DNA loss
- Sensitive: Works with as low as 5000 cells
- Fast: Sample to single-cell platform in 1 day
- Flexible:: Integration with existing single-cell platforms
3. Scalability to Tens of Thousands of Cells
By coupling scCUT&Tag with droplet or nanowell barcoding systems, researchers can profile tens of thousands of cells in a single experiment. Combinatorial indexing and high-throughput implementations further increase scalability, enabling cost-effective mapping across large cell populations and experimental conditions.
- 10x Chromium: 10K-20K cells per reaction
- BD Rhapsody: 50K-80K cells per reaction
- Illumina PIPSeq: 2K-100K cells per reaction
- High-Complexity: Deep per-cell coverage for robust peak calling
4. Direct Regulatory Insights and Rare Cell Detection
scCUT&Tag simultaneously captures active and repressive histone marks and transcription factor occupancy, enabling direct inference of promoter activity, enhancer usage, bivalency, and long-range regulatory features. The method reliably recovers major cell types and resolves rare or transitional populations invisible to bulk assays.
- Regulation: Cell-type-specific histone mark and TF landscapes
- Cell transitions: Detection of rare and transitional cell states
- Network biology: Direct regulatory network mapping in specific cell populations
- Chromatin states: Bivalent mark identification for developmental priming
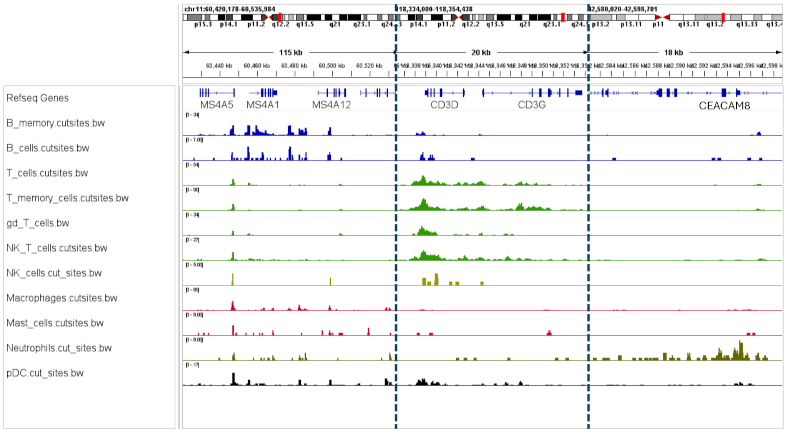
Research Applications
Developmental Biology
Map chromatin remodeling and histone modification changes during differentiation and cell-state transitions. Identify epigenetic priming events and cell-fate decisions.
- Cell-fate specification mechanisms
- Epigenetic priming during lineage commitment
- Bivalent mark identification at developmental genes
Cancer and Tumor Heterogeneity
Characterize intra-tumoral epigenetic heterogeneity, identify therapeutic resistance drivers, and map immune infiltration states via chromatin profiling.
- Tumor-associated macrophage epigenetics
- T cell exhaustion and activation states
- Cancer cell clonal heterogeneity
Immunology and Cell State Dynamics
Profile immune cell heterogeneity and activation states without requiring transcriptomics. Map histone marks and TF binding during immune responses.
- T cell exhaustion and memory programs
- B cell differentiation and plasma cell states
- Myeloid cell activation states
Perturbation Studies & Drug Response
Measure chromatin changes following CRISPR, compound treatments, or genetic perturbations. Map epigenetic responses without requiring transcript quantification.
- CRISPR screen validation via chromatin changes
- Drug response epigenetic signatures
- Chromatin remodeler targeting and validation
Single-Cell CUT&Tag vs. Alternatives
| Aspect | scCUT&Tag | scATAC-seq | ChIP |
|---|---|---|---|
| Primary Readout | Specific histone marks / TF binding | General chromatin accessibility | Specific marks, often lower sensitivity |
| Sensitivity & Noise | High sensitivity, low background | Good sensitivity, more off-target signal | Often lower signal-to-noise |
| Input & Depth Required | Very low cell input, low sequencing depth | Moderate input, higher depth for coverage | Typically higher input and depth |
| Throughput & Scalability | Tens of thousands of cells per run | Tens of thousands of cells per run | Bulk assay - not single-cell |
| Type of Insights | Direct chromatin mark and TF occupancy | Global accessibility landscapes | Chromatin mark and TF occupancy |
When to Choose scCUT&Tag
- Focused histone profiling: When specific marks (H3K27ac, H3K4me1, H3K27me3, etc.) or TFs are your primary interest
- Limited sample availability: Ultra-low-input capability ideal for precious tissues or clinical samples
- Archived or compromised samples: FFPE tissues or degraded RNA where RNA-based methods fail
- Large-scale profiling: Combinatorial indexing enables 100K+ cell experiments cost-effectively
Integration with Multiomic Assays
scCUT&Tag integrates seamlessly with emerging multiomic workflows that combine chromatin profiling with transcriptomics or multiplexed measurements, enabling coordinated analysis of gene expression and chromatin state in the same single cells. Long-read and spatial implementations extend chromatin profiling into repetitive regions while preserving tissue context.
Advanced Single-Cell Chromatin Profiling
Single-cell CUT&Tag revolutionizes epigenetic research by delivering sensitive, targeted chromatin profiling at single-cell resolution with minimal input and cost. Whether you're mapping developmental epigenetics, characterizing tumor heterogeneity, or studying immune cell states, scCUT&Tag provides the resolution and scalability needed to unlock mechanistic insights into cellular identity and regulation.